Jupyter notebooks are a constant source of debate in the data science community. Should they be used for prototyping, in production, as an end-to-end development environment for creating applications and packages or not at all? Here are three principles I found apply irrespective of how you are using notebooks.
1. Make the setup work for you
Getting started with Jupyter is easy: Install jupyterlab or notebook locally, and you’re good to go. Also, cloud providers have a variety of managed notebook offerings, some part of their ML platforms, (e.g., AWS Sagemaker, Azure Machine Learning Studio Notebooks, GCP AI Platform Notebooks, Google Colab, …) that can get you started quickly. While the first steps are usually simple, there are a few things to consider when choosing a setup.
Dependency management
Python and dependency management is a complicated relationship to say the least. But whether you’re using conda, venv, pipenv, virtualenv, poetry, or whatever the latest tool is - make sure you have a way to link your notebook with a reproducible environment specification. I usually install Jupyter into my base conda environment and use venv or conda virtual environments for project dependencies specified in requirements.txt or environment.yml files. I then link project-specific environments to my main Jupyter installation by setting up an ipykernel (see the ipython docs for details). This setup allows me to manage general configurations (see the section on customizability) centrally while separating project dependencies cleanly. It also makes it easy to install custom applications still under development and neatly integrate module code with exploratory work in notebooks. See 2. Write clean code for more details.
Transparency and customizability
I have a conflicting relationship with managed notebook offerings in the cloud like AWS Sagemaker or Azure Machine Learning Studio Notebooks. On the one hand, they make getting started easier and facilitate integration with other cloud services. On the other hand, I found the setup happening behind the scenes can be quite opaque and hard to customize. Why is customizability important to me? In my experience, the more productive engineers are, the happier they feel. Others wrote about how improvements in engineers’ productivity translate into a competitive advantage and keep the positive flywheel for software engineering as a whole going. I know that tying the customizability of Jupyter notebooks to competitive advantages is a stretch. My point is that sacrificing it at the expense of simplicity and standardization is a trade-off worth considering.
When I say “customizability”, I primarily mean how easy it is to configure tools that simplify my life as a developer. For me, automatically formatting notebook code using Black is a tool that I would only reluctantly give up, for example. Here’s a Dockerfile extending jupyter/base-notebook that illustrates the configuration I like to use. Another interesting approach to customizing notebooks is using Jupyter as Python source files in VS Code. This approach looks promising to me as it allows for using the IDE configuration while working in a notebook-like interactive environment. Regardless of which tools you use: Ask yourself regularly if they support you in being as productive as you could be.
Shared datastore
Cookiecutter Data Science, a popular template project structure for data science projects, makes a convincing case for organizing raw input data, intermediate / transformed data, and processed results data separately in a local data directory. I wholeheartedly agree with the notion that raw data should be immutable, and layers separating raw, intermediate, and results data are valuable to keep things organized. However, when collaborating in teams larger than ~2, I found establishing a standard local storage structure difficult and felt that this approach quickly causes redundancies. Therefore, whenever possible, I stream data from a remote store accessible by everyone on the team (e.g., object storage such as S3 or Azure Blob) and avoid local files. The layer architecture (raw, intermediate, results, or similar) can be enhanced with layer-specific access rules (e.g., read-only for raw data) and allows for sharing common data in the raw and intermediate layer between projects. Only working with data in central remote storage means more effort in the beginning (e.g., pd.read_csv works with local file systems, but not with all remote stores out of the box and remote stores require secrets handling), but usually pays off quickly by improving collaboration and facilitating reproducibility.
2. Write clean code
While the setup is important, what’s in the notebook arguably matters even more. In his presentation titled “I don’t like notebooks” at JupyterCon 2018, Joel Grus argued that notebooks encourage writing messy code (I am paraphrasing here, see his slides for details).
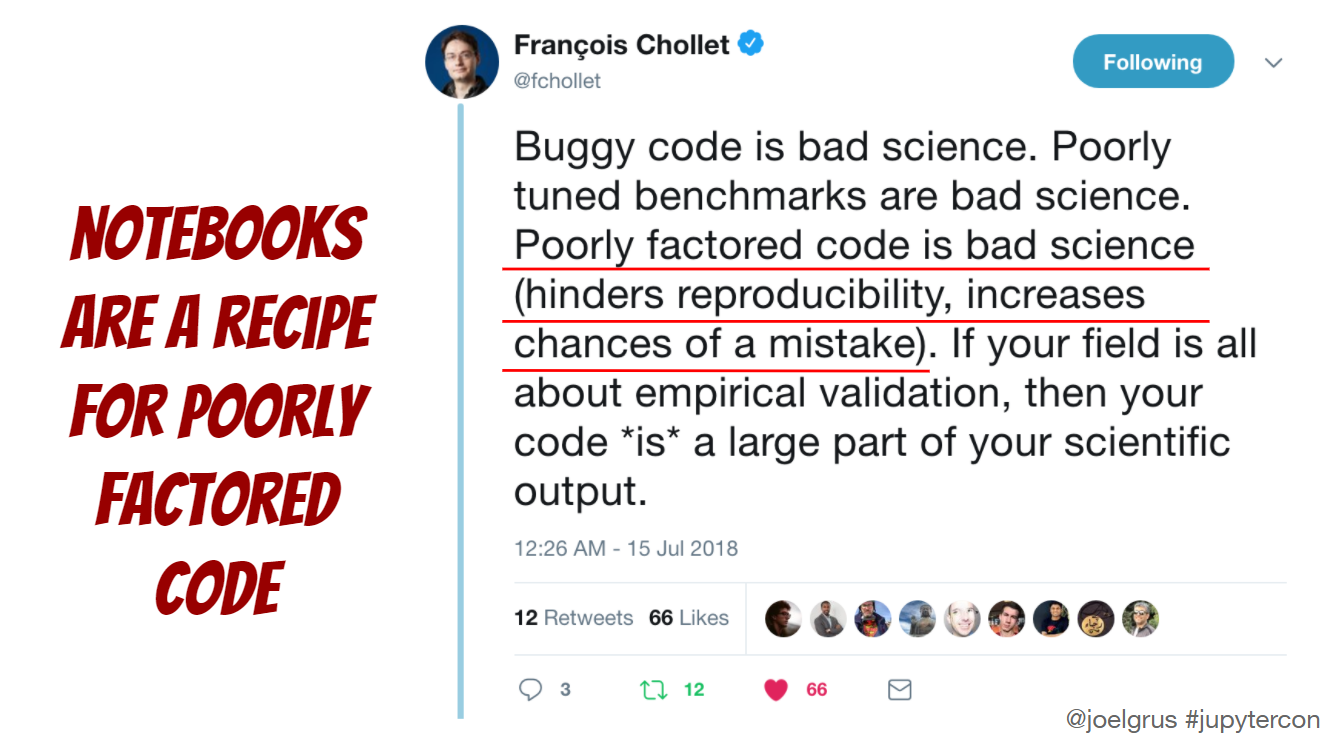
I see the point and have dealt with my fair share of messy notebook code (and produced more such code than I would like to admit). But I have also seen several projects using notebook and module code (in .py files, potentially organized as a package) in a way that played to their respective strengths. Notebooks for experiments, prototypical implementation, analyses, or reports leveraging their interactivity and ability to integrate code, markdown, and output. Module code for reusable, tested artifacts intended for sharing with others or your future self building on top of an established tool landscape for testing, linting, automatic code formatting, etc. in an IDE.
fastai’s nbdev attempts to unify these strengths using notebooks as the main development environment. I find this approach intriguing but haven’t tried it on a serious project yet.
Irrespective of your favorite development environment, tools, and processes: Making it easy to factor out reusable functionality from notebook to module code early and regularly (avoiding the worst-case - excessive copy-pasting between notebooks) is key for reproducibility, maintainability, and ultimately, staying sane. The details of how to achieve this go beyond the scope of this post. Have a look at the project template for Python apps I like to use that includes helpful functionality and instructions encouraging such a workflow (disclaimer: I’m a maintainer of the template).
One additional, concrete technique I found to make my notebooks cleaner and more readable is using method chaining when writing data transformation code in Pandas. I first learned about method chaining from Tom Augspurger’s blog series on Modern Pandas (almost five years old at the time of this writing, but still full of great insights) and found it immediately appealing. It improves readability and encourages writing pure functions from DataFrame –> DataFrame that implement atomic transformation steps. This makes unit testing easier - debugging becomes harder though for long transformation chains since it’s harder to inspect intermediate results. Method chaining is the Pandas equivalent to pipes in R’s tidyverse (%>%) and the functional style of Spark’s DataFrame API.
3. Have one purpose
For my last principle, let’s revisit the slides from Joel’s JupyterCon talk.

Here, Joel makes the point that hidden state makes notebooks hard to reason about (in this case due to a cell that was deleted after it was executed), even more so “with dozens of cells and more complex code”. He goes on to say that “out-of-order execution makes learning Python more confusing than it needs to be” for beginners. In my experience, the general point about hard-to-reason-about notebooks applies beyond beginners learning Python.
It can be tempting to keep adding “small experiments” to your notebook, jumping back and forth between different parts “quickly changing” previous steps to test something, and never taking the time to clean up. That’s how long, convoluted notebooks develop and become difficult to understand for others or your future self.
For me, the key to avoiding such a mess is to focus, i.e., have one clearly defined purpose: An analysis to be shared and discussed with a business stakeholder, an experiment evaluating an algorithm from the literature on our data, or a prototypical implementation of a new metric - whatever it is, stay focused and keep the notebook as short as possible. By any means, try that idea for an additional plot or alternative metric definition. But don’t hesitate to remove obsolete cells or split one notebook into multiple to avoid a long, cluttered result combining too many ideas, experiments, or analyses. I learned this the hard way: Too often have I thought “this might become relevant at some point” and created notebooks I wasn’t very excited about revisiting a few months down the road. You probably guessed it: Never has “this” become relevant again…
Keeping notebooks short and focused brings you a long way. There’s one other important rule to avoid creating a mess: Making sure the notebook can be executed from top to bottom. Most of the time, I just restart the kernel and re-run everything before committing my work. If executing everything takes way too long, you might want to reconsider whether your notebook is short / focused enough or evaluate whether you should create an intermediate data set, factoring out data preprocessing from your analysis.
Summary
There you go. These are the three principles that help me make the most of Jupyter notebooks:
- Optimize my setup to foster collaboration with my team, support reproducibility, and make my life as a developer as easy as possible.
- Don’t use notebooks as an excuse for writing messy code. To avoid messy code, I factor out reusable pieces from the notebook into tested module code early and regularly.
- Keep notebooks short and focused on a single purpose and ensure they run from top to bottom.
One last thing: I’m convinced that there is no one correct way of developing software or doing data science. There are good and bad practices. There are better and worse tools. What’s important is to regularly consider whether your practices and tools optimally support your objectives - such as collaboration, reproducibility, developer productivity, and not making a mess.